Protein folding and protein interactions underlie both proper function and disease in many biological systems. Many receptor proteins signal through complex interactions and rearrangements, such as the SOS signaling pathway that governs antibiotic resistance in bacteria, and some proteins, such as the Parkinson’s Disease protein α-synuclein, misfold into toxic conformations. Studying these protein motions not only aids our understanding of diverse biological phenomena, it also contributes to an important fundamental problem in biochemistry: understanding how proteins fold and change shape. The Petersson laboratory is developing tools to address questions of how peptides and proteins mediate cellular communication and how the cellular environment catalyzes protein misfolding, from detailed in vitro folding studies, to tracking protein aggregation in neurons, and even to human clinical imaging. These tools include novel chromophores, which we synthesize and incorporate into proteins through unnatural amino acid mutagenesis and synthetic protein ligation. In many cases, protein misfolding is regulated by post-translational modifications, for which we study the enzymes that install them, both to understand their biological roles and to utilize them in synthetic protein modification. Finally, an area of particular interest in the Petersson laboratory is the introduction of thioamide modifications to the peptide backbone, which can serve as protein folding probes, or stabilizers for improved therapeutic peptides or in vivo imaging reagents.
| |
|
|
|
|
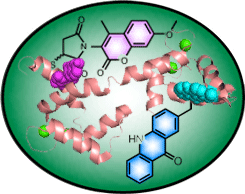
Minimal Chromophores to Monitor Protein Motions
Fluorescence and FRET measurements can be used to glean time-resolved structural information on protein motions. However, the relatively large size of common fluorophores can lead them to disrupt the very process they are meant to report on. We have developed fluorescent amino acids and reagents that can be used to label a protein without disrupting its fold or function. |
| |
|
|
|
|
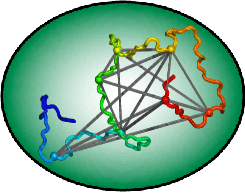
Computational Modeling of Dynamic Protein Structures
In order to acheive structural models of disordered proteins and protein conformational motions, we use distance data from FRET and other measurements to drive Monte Carlo simulations. We combine these data with high resolutuon X-ray, cryo-EM, or solid state NMR data to make models of complex, dynamic proteins that no one technique can visualize. |
| |
|
|

|
|
Toward a Molecular Understanding of Parkinson's Disease
Parkinson's Disease is one of the leading causes of death in the United States, yet its etiology remains unknown. Using chemical biology tools, we study the molecular basis for the disease, both through in vitro stduies of the misfolding of purified proteins like alpha-synuclein and tau, through microscopy studies that allow us to observe the affects of aggregated proteins in cells, and even through in vivo imaging (in collaboration with radiology groups at Penn and elsewhere). |
| |
|
|
|
|
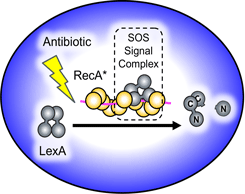
Targeting the Mechanisms of Antibiotic Resistance
Antibiotic resistant bacterial pathogens pose a major public health threat. In collaboration with the Kohli laboratory in the Department of medicine, we use a combination of chemical, biochemical, microbiological, and computational approaches to find vulnerabilities in the SOS pathway, a critical genetic response networklinked to bacterial adaptation and evolution. |
| |
|
|
| |
|
|
|
|
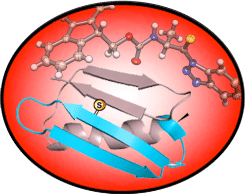
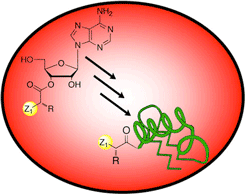
Thioamide Modifications of the Protein Backbone
Thioamides have subtly different steric and electronic properties than oxoamides that allow them to precisely disrupt protein interactions, or act as site-specific photoswitches or fluorescence quenchers. We have shown that thioamides can be used to monitor protein folding and proteolysis using fluorescence, and that thioamide-modified peptides can be stable and bioactive.
|
| |
|
|
|
|
Post-Translational Modification Enzymology and Applications
Post-translational modifications are ubiquitous and govern many Important cellular processes such as protein-protein interactions and degradation. We study the molecular basis for these modifications using semi-synthesis as a tool for site-specifically manipulating proteins and attaching functional probes.
|
| |
|
|
| |
|
|
| |
|
|
| |
|
|
| |
|
We are currently funded by the following agencies: |
| |
|
|
 |
|
The National Science Foundation (CHE-2203909)
|
 |
|
The Department of Energy (DOE-SC0022240) |
 |
|
The National Institutes of Health
(RF-1NS103873, RF1NS125770, R01-GM127593, R01-NS102435, and U19-NS110456)
|








